DimerDice allows visualizing different supramolecular dimeric complexes of cyclic peptides in Augmented Reality (AR)
Dimer dice

What is it about?
DimerDice allows visualizing different supramolecular dimeric complexes of cyclic peptides in Augmented Reality (AR). The cyclic peptides (CPs) are composed by γ-amino acids (cis-α,γ-aminocycloalkanecarboxylic acids, γ-Acas) alternated with α-amino acids (α,γ-CPs). These peptides containing γ-Acas can be designed from the original D,L-α-CPs just by replacing any α-amino acid for a γ-Aca of equivalent chirality. For instance, L-amino acids can be replaced by (1R,3S)-isomers while the (1S,3R)-Acas would substitute the D-enantiomers.

App Store Description
DimerDice allows visualizing different supramolecular dimeric complexes of cyclic peptides in Augmented Reality (AR). The cyclic peptides (CPs) are composed by γ-amino acids (cis-α,γ-aminocycloalkanecarboxylic acids, γ-Acas) alternated with α-amino acids (α,γ-CPs). These peptides containing γ-Acas can be designed from the original D,L-α-CPs just by replacing any α-amino acid for a γ-Aca of equivalent chirality. For instance, L-amino acids can be replaced by (1R,3S)-isomers while the (1S,3R)-Acas would substitute the D-enantiomers.
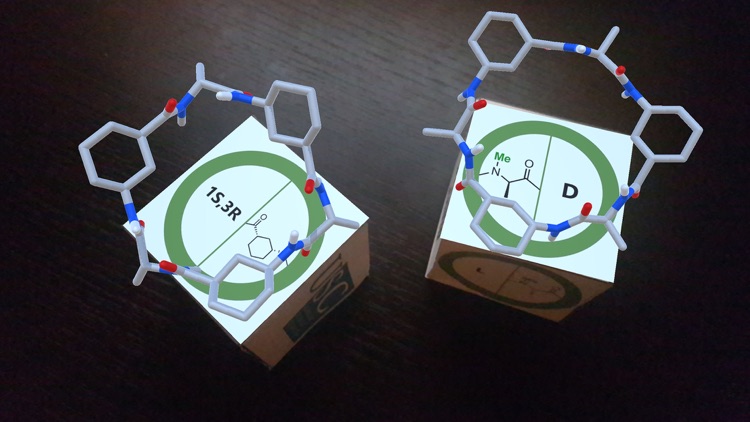
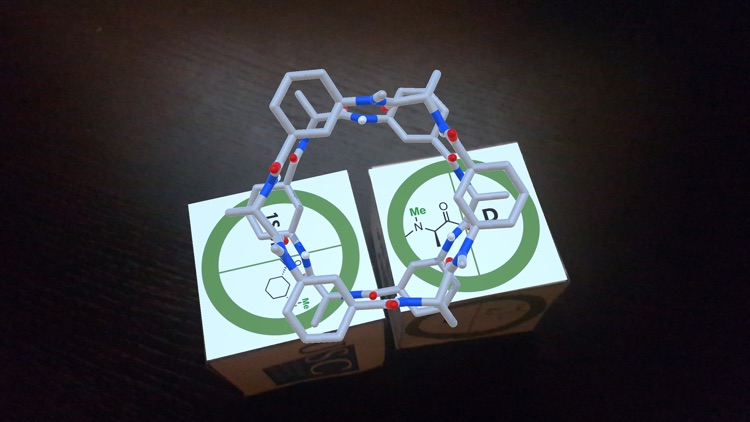
As part of an in-vitro/in-silico study carried out at Santiago de Compostela University to deeply analyze the preferred rearrangement adopted by α,γ-CPs, we explored all the possible combinations that could take place starting from two different monomers and their corresponding enantiomers.[1] This approach uses the selective N-methylation of the amide groups whose protons are not involved in hydrogen bonding interactions. The N-methylated residues are indicated in the figures of the trackers. Depending on the combination of monomers, a parallel or an antiparallel arrangement is possible.
Pointing with the device to each tracker separately, an AR image of the corresponding monomer is viewed. When the tracker images of two monomers are placed close to each other, the structure of the dimer formed by that combination of cyclic peptides is shown.
Full geometry optimizations of the dimers and monomers were carried out with the B3LYP functional and the standard 6-31G(d) basis set. All of the DFT calculations reported in this study were performed with the Gaussian 16 package.
This application was specifically developed by Santiago de Compostela University (Spain) and MD.USE Innovative Solutions S.L. (www.mduse.com).
[1] Manuscript in preparation.
AppAdvice does not own this application and only provides images and links contained in the iTunes Search API, to help our users find the best apps to download. If you are the developer of this app and would like your information removed, please send a request to takedown@appadvice.com and your information will be removed.